What if the defining reason for life on Earth is the manipulation of time? While organic matter has been discovered in outer space, in the form of methane in the ethers of Neptune and Uranus, and asteroids and meteorites that crashed into Earth (Organic Compounds in the Solar System, 2016), why have we not yet discovered extraterrestrial life in the form of a cell that is moving, eating, growing, changing, and replicating (Sagan & Sagan, 2018)?
The defining feature of life could be the enzyme: proteins made from amino acids that increase the rate of chemical reactions. Metabolic reactions could not occur without enzymes (Lewis & Stone, 2020), as reactions would occur too slowly to interact with one another. Enzymes increase the speed of reactions so that complex, multifaceted interactions can occur to create and sustain life. It is the enzyme that has catalyzed life on Earth, by shortening the time needed for reactions to take place.
Although amino acids have been discovered in some meteorites, such as the simpler alanine and glycine found in class CI and CN meteorites (Organic Compounds in the Solar System, 2016), the 20 main ones that define life on Earth have not yet been found elsewhere in the universe in same concentrations and varieties.
There is much debate surrounding the origin of the 20 amino acids. Is it by chance or by evolutionary design? And are more amino acids evolving or could they evolve? Other amino acids do exist, including selenocysteine which is found in humans, but is comparatively more complex to utilize, which might be a clue as to why there aren’t more than the 20 main ones that exist (Brazil, 2017).
Francis Crick hypothesized in his frozen accident theory that 20 specific amino acids exist by chance and that any other number of structures could have been used to synthesize proteins (Koonin, 2017). However, in 2017 Doig makes strong arguments that the 20 main amino acids with their specific hydrophobicity enable folding, stability, accessibility of active sites, and metabolic efficiency, making amino acids the perfect building blocks for proteins and enzymes (Doig, 2017).
Why are C, H, N, O, and S the organic molecules that compose amino acids? In addition to being bountiful on our planet, these elements are ideal for many reasons. In contrast, metals such as selenium and antimony are too soluble in water and therefore unstable. Halogens are too electronegative and reactive. Silicon bonds too readily to oxygen in place of other elements. The list goes on and on to account for the specificity that explains why our current amino acids are the perfect constituents of life (Doig, 2017).
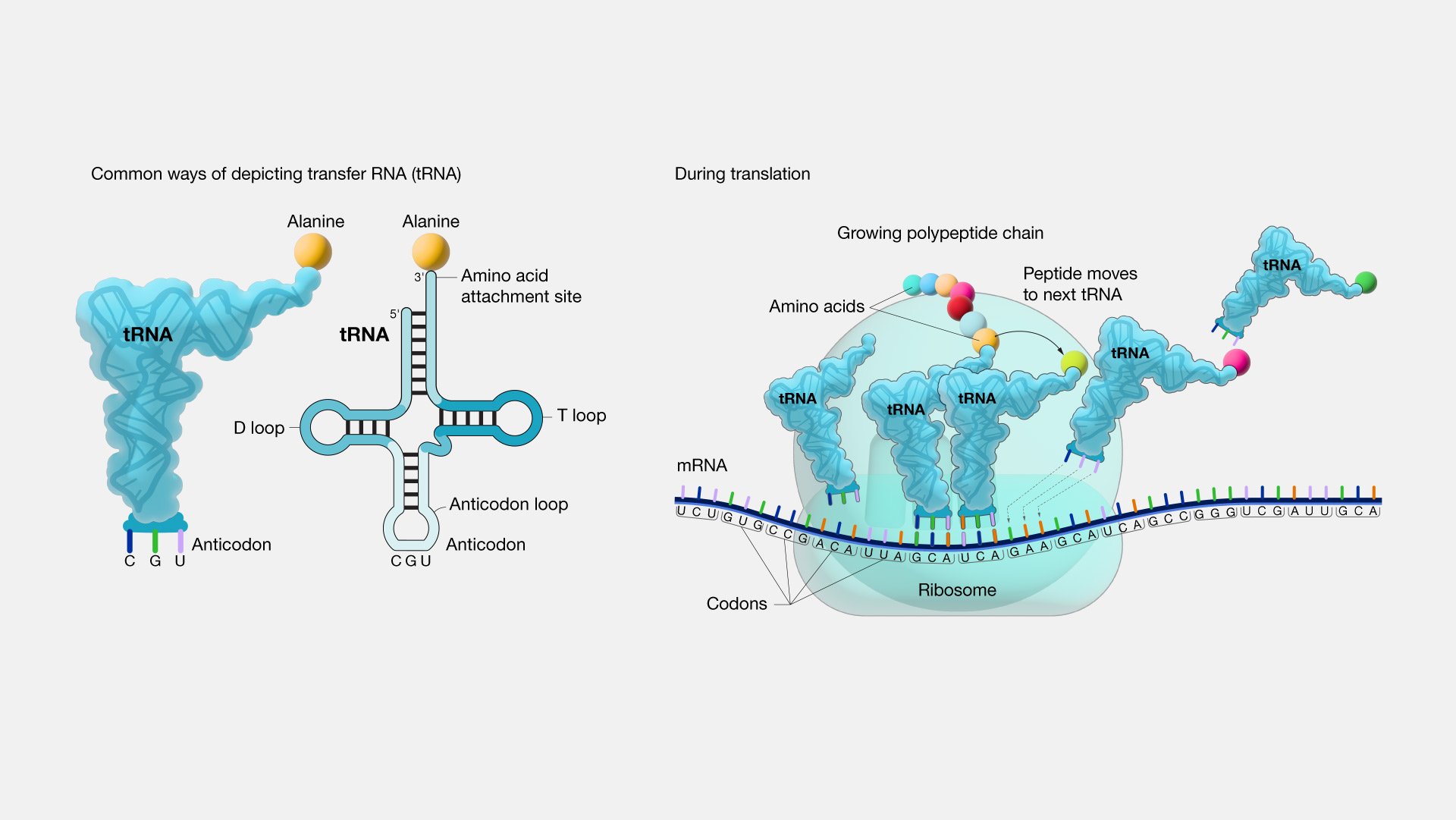
So, if it is not a frozen accident then why has the number of amino acids stopped at approximately 20 when so much of evolution is characterized by seemingly endless diversity? One fascinating theory explains it in terms of simple logistics. tRNA (transfer RNA) reads the genetic code from RNA, and then using the enzyme aminoacyl tRNA synthetase, selects which amino acid tRNA should bind to. tRNA then transfers the selected amino acid to a ribosome for assembly on the growing polypeptide chain. tRNA has only 3 reading sites, and after accounting for the start and stop codons, 61 possible amino acids reading codes. However, tRNAs are limited in their recognition ability (Saint-Léger et al., 2016). tRNAs already make on average one mistake for every 1000-10,000 codon readings and adding more amino acids would enable more mistakes. Dr. Ribas explains it this way, “It’s like if you have a very simple kind of lock where you could only change three or four pins, you come to a point where you wouldn’t be able to make new keys because a new key will open a lock you have already used and that defeats the purpose” (Brazil, 2017).
Photo Credit: (National Human Genome Research Institute, 2019)
Scientists do not have all the answers on whether amino acids evolved by chance or limited necessity, but one thing is certain; the 20 amino acids are brilliant in their specificity and enable life as we know it. Without proteins, we would not have enzymes, and without enzymes, we would still be a pool of RNA and co-factors. Enzymes alter the speed of reactions and amino acids are the specific building blocks of all proteins and enzymes. Without our 20 amino acids, we would lack the complex web of reactions needed to create life; organisms that can move, eat, breathe and reproduce.
REFERENCES
Organic Compounds in The Solar System. (2016, May 26). Chemistry LibreTexts. https://chem.libretexts.org/Ancillary_Materials/Exemplars_and_Case_Studies/Exemplars/Physics_and_Astronomy/Organic_Compounds_in_The_Solar_SystemLinks to an external site.
Sagan, D., & Sagan, C. (2018). life | Definition. In Encyclopædia Britannica. Retrieved from https://www.britannica.com/science/lifeLinks to an external site.
Lewis, T., & Stone, W. L. (2020). Biochemistry, Proteins Enzymes. PubMed; StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK554481/#:~:text=Enzymes%20are%20proteins%20comprised%20ofLinks to an external site.
Brazil, R. (2017). Why are there 20 amino acids? Chemistry World. https://www.chemistryworld.com/features/why-are-there-20-amino-acids/3009378.articleLinks to an external site.
Doig, A. J. (2017). Frozen, but no accident – why the 20 standard amino acids were selected. The FEBS Journal, 284(9), 1296–1305. https://doi.org/10.1111/febs.13982 Links to an external site.
Koonin, E. (2017). Frozen Accident Pushing 50: Stereochemistry, Expansion, and Chance in the Evolution of the Genetic Code. Life, 7(2), 22. https://doi.org/10.3390/life7020022 Links to an external site.
Saint-Léger, A., Bello, C., Dans, P. D., Torres, A. G., Novoa, E. M., Camacho, N., Orozco, M., Kondrashov, F. A., & Ribas de Pouplana, L. (2016). Saturation of recognition elements blocks evolution of new tRNA identities. Science Advances, 2(4). https://doi.org/10.1126/sciadv.1501860Links to an external site.
National Human Genome Research Institute. (2019). Transfer RNA (tRNA). Genome.gov. https://www.genome.gov/genetics-glossary/Transfer-RNA